Three years ago, I gently brushed fibre-tipped swabs against the surfaces of my tiny apartment. Microbes live everywhere, and I was gathering samples for genetic analysis — I wanted to identify my microscopic housemates.
Just as microbes live all over our bodies, they also live all over our pillows and furniture and doorknobs. But we still know very little about them.
Last month, a massive study sequencing all the lifeforms on the New York City subway found that nearly half of the DNA matched no known organism. And the DNA matches that researchers did get included bubonic plague, Tasmanian devil and Himalayan yak. What?!
Amidst all the confusion and subway plague panic, I decided to take a closer look at the New York City microbial community I knew most intimately — that of my former apartment. Back in 2012, I sent microbial samples from my apartment to a citizen science project called The Wild Life of Our Homes, run out of North Carolina State University. The researchers there promised to sequence the DNA from what I’d sent them.
About six months ago, I finally got the the first results: two pie charts and an Excel spreadsheet full of names I couldn’t really pronounce and definitely couldn’t spell. (
Corynebacteriaceae and
Sphingomonadaceae, to name a couple).
As I attempted to understand the data that summed up many of the invisible life forms that lurked in my apartment, I came up against the limits of modern science. The fact is, we still don’t understand much about microbiomes, or ecosystems of microbes. And the more you know, the more realise you don’t know anything.
Is This Bacteria Really From Poop?
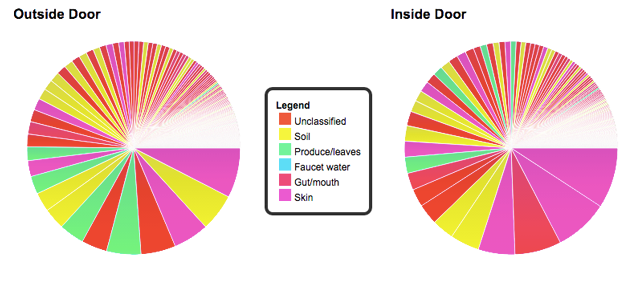
The bacteria found in my apartment. I know, the colours of “unclassified” and “gut/mouth” bacteria in the pie chart above are very, very similar. Sorry! I didn’t make the charts! If you want to know more about all the unclassified bacteria, here’s the entire raw spreadsheet. Credit: The Wild Life of Our Homes.
After I got my pie chart and spreadsheet, I called up Rob Dunn, a biologist at North Carolina State University and the guy behind Wild Life of Our Homes, to talk through my results in more detail. In sampling my home, I had gently swabbed four places: pillow, kitchen counter, outside doorframe, and inside doorframe. Only the results of my doorframe were available so far.
One of the most common bacteria on the inside of my doorframe was
Sphingomonadaceae, a soil microbe that had perhaps hitched a ride on the soles of my shoes. The outside door frame was dominated by Micrococcaceae, a bacteria that likely flaked off with my skin. Dunn also pointed out Corynebacteriaceae, which usually lives in the sweaty swamp of our armpits.
I went further down the list in search of more exotic microbes. One bacterial family unusually common in my apartment was
Deinococcaceae, classified under produce/leaves. There’s not much we know about this microbe, but I did dig up one paper that found a species of it living in citrus leaf canker lesions, which made me wonder about all the grapefruits I used to buy in Chinatown.
But even as I was trying to make sense of the categories of bacteria in my former home — and with a slightly triumphant note of see mum, my first post-college apartment wasn’t that filthy — uncertainty plagued me. “Most of the microbes in your home are too poorly known to even guess where they might be coming from,” read the statement about my pie chart results. “And even among well-studied organisms, we have uncertainty.”
In fact, sorting bacteria into categories based on where they come from, like skin, faeces and soil, is really just our feeble human minds imposing artificial order on a natural world. A type of bacteria, Lactobacillaceae, might become known as faecal bacteria because we find a lot of it in the human gut, but that doesn’t mean it grows there exclusively. Just because there is Lactobacillacae on my door frame (which, for the record, there was) doesn’t mean it got there through a smear of poop.
Jonathan Eisen, a microbiologist at the University of California, Davis, has an analogy for why these categories can be misleading. There are a lot of rats in New York City, but “finding a rat in your backyard does not mean you live in New York City.” I, at least, am choosing not to freak out about the so-called faecal bacteria in my apartment.
We Can’t Identify A Lot of the Bacteria We Do Find
A total of 965 bacterial families were found on the inside and outside frame of my apartment door. It’s hard to see laid out in the pie chart, but there is a very long tail and most of that is “unclassified bacteria.”
If you peer at the microbial long tail, you start to see a lot of microbes that have a kingdom, phylum, and class classification, but that’s it. (That means they haven’t even been classified into order, family, genus, and species.) Dunn told me these unclassified bacteria contain DNA sequences that don’t match those from any named and described microbes.
In fact, the vast majority of bacteria that researchers have described are the ones that are easily grown in a petri dish. Depending on who you ask, that represents 1 to 5 per cent of all bacteria out there, possibly even less.
This problem of how much we don’t know is especially acute with a relatively new technique called metagenomic sequencing that’s become popular as DNA sequencing has become cheaper. With metagenomic sequencing, you sequence an entire microbiome, or collection of microbes from a single location. That means the sequence results are a hodgepodge of hundreds or even thousands of different organisms.
The bacteria in my apartment were identified by sequencing only a portion of a gene called 16S rRNA, a traditional technique that help researchers pinpoint bacteria at the family or genus level.
Metagenomics is potentially much more powerful because it sequences entire genomes. Once all the genetic material in a sample has been sequenced, there are software programs that can match up the DNA segments with bacterial genomes in data libraries.
Of course that assumes that the DNA you’ve sequenced is even in those libraries. There are 100 or so major lineages of bacteria, and the vast majority of bacterial genomes we’ve sequenced come from just three. We have moderate sampling of genomes from about 10 other linages. “And the remaining 80 we suck at,” says Eisen.
Suppose here’s a bacteria called
Gizmodoacae out there. If we don’t already know
Gizmodoacae‘s genome sequence — and chances are, we don’t — we can’t know that we found Gizmodoacae in our sample. Our poor bacteria falls into the cracks of our knowledge.
We Find Stuff (Plague, Platypuses, Yaks) That Doesn’t Make Sense
Perhaps even more troubling is that these analyses can sometimes “find” bacteria that don’t actually exist in a sample. This is a particular problem with metagenomics sequencing, which involves chopping up a whole bunch of genomes and piecing them back together. Sometimes the software prgorams that do the piecing together can go very wrong.
For example, the recent subway microbiology study “found” DNA sequences that plainly didn’t make sense: Tasmanian devil, Himalayan yak, and the Mediterranean fruit fly. The researchers threw these out. They did, however, mention finding possible evidence for plague bacteria on the subway.
The plague mention sparked a lot of headlines but also quite a bit of criticism. In a blog post several days after, the research team explained in gory technical detail how they got the hit for plague bacteria. Here’s the gist: Many lethal bacteria are closely related to harmless ones, like the many different strains of E. coli. It was likely they had found a harmless relative of the plague bacteria, but their reference libraries contained only the genome of the harmful one. Hence, a hit for plague.

Dear platypus, what is your DNA doing on tomato plants in Virginia? worldswildlifewonders/shutterstock
Plenty of other examples abound, many of them collected by Ed Yong in an excellent piece for his blog, Not Exactly Rocket Science. Platypus has been found on tomato plants in Virginia. Nick Loman of the University of Birmingham the genome sequence forEscherichia coli, chopped it up into 100 base pair segments, and put it through a typical algorithm for metagenomic analysis. He obviously should have ended up with 100 per cent E. coli, but the algorithm spit out 61.3 per cent.
Our data are only as good as the tools we use to interpret them. There are ways to get around these false positives, but they require more computational power.
The Future Beckons
The field of microbiology today is roughly where all of biology was at when Darwin set sail on the Beagle and began gathering evidence for evolution by meticulously studying species variation on islands. We’re in the cataloguing phase. That means we’re going to have to be patient while scientists gather data. But that also means we’re on the verge of something big.
The microbiome of my apartment alone doesn’t tell scientists — or even me — very much. But it is part of a much larger citizen science project comparing the microbiomes of nearly 1,500 houses all over the country. And to get past the cataloguing phase, scientists have to do experiments. What happens to a house’s microbial community when you introduce a dog? Or triclosan, an antimicrobial commonly found in household products? There are many stories left to tell.
“You can walk through your house and hold our your hands, and there are these mysterious cells,” says Dunn. “Each one of those cells is the tip of a thread of a big rope of a story. Those tips are all around is waiting around us for us to unravel. Just think of the richness of life that we have yet unravel. ”
I have to agree with him.
Picture: Staphylococcus aureus. Janice Haney Car/CDC